Code
fit_verb_fp_mm <- lmer(log(fp) ~ verb_t*gramm +
(1 + verb_t*gramm|sj) +
(1 + verb_t*gramm|item),
data = df_biondo,
subset = roi == 4)Strolling through the garden of forking paths
Daniela Palleschi
Humboldt-Universität zu Berlin
February 9, 2024
March 11, 2024
This chapter is not fully translated from bullet points (from my slides) to prose. This will happen eventually (hopefully by spring 2024).
Today we will learn about…
and how to…
A maximal model should optimize generalization of the findings to new subjects and new items.
– Barr et al. (2013), p. 261
[W]hile the maximal model indeed performs well as far as Type I error rates were concerned, power decreases substantially with model complexity.
— Matuschek et al. (2017), p. 310-311
Every statistical model is a description of some real or hypothetical state of affairs in the world.
– Yarkoni (2022), p. 2
# A tibble: 1 × 2
n nn
<int> <int>
1 16 240What we hope to make clear is that there is no single correct way in which LMM analyses should be conducted, and this has important implications for how the reporting of LMMs should be approached.
— Meteyard & Davies (2020), p. 9
Replicability and reproducibility are critical for scientific progress, so the way in which researchers have implemented LMM analysis must be entirely transparent. We also hope that the sharing of analysis code and data becomes widespread, enabling the periodic re-analysis of raw data over multiple experiments as studies accumulate over time.
— Meteyard & Davies (2020), p. 9
We’ll now look at an example of a model that encounters convergence issues, and take some steps to reach convergence. We are considering a case where your predictors and covariates are already selected, and will focus on selecting model options and random effects structure that achieve model convergence.
We first need to set up our environment.
df_biondo <-
read_csv(here("data", "Biondo.Soilemezidi.Mancini_dataset_ET.csv"),
locale = locale(encoding = "Latin1") ### for special characters in Spanish
) |>
clean_names() |>
mutate(gramm = ifelse(gramm == "0", "ungramm", "gramm")) |>
mutate_if(is.character,as_factor) |> ## all character variables as factors
droplevels() |>
filter(adv_type == "Deic")nloptwrap from bobyqa, there seem to be more ‘false positive’ convergence warnings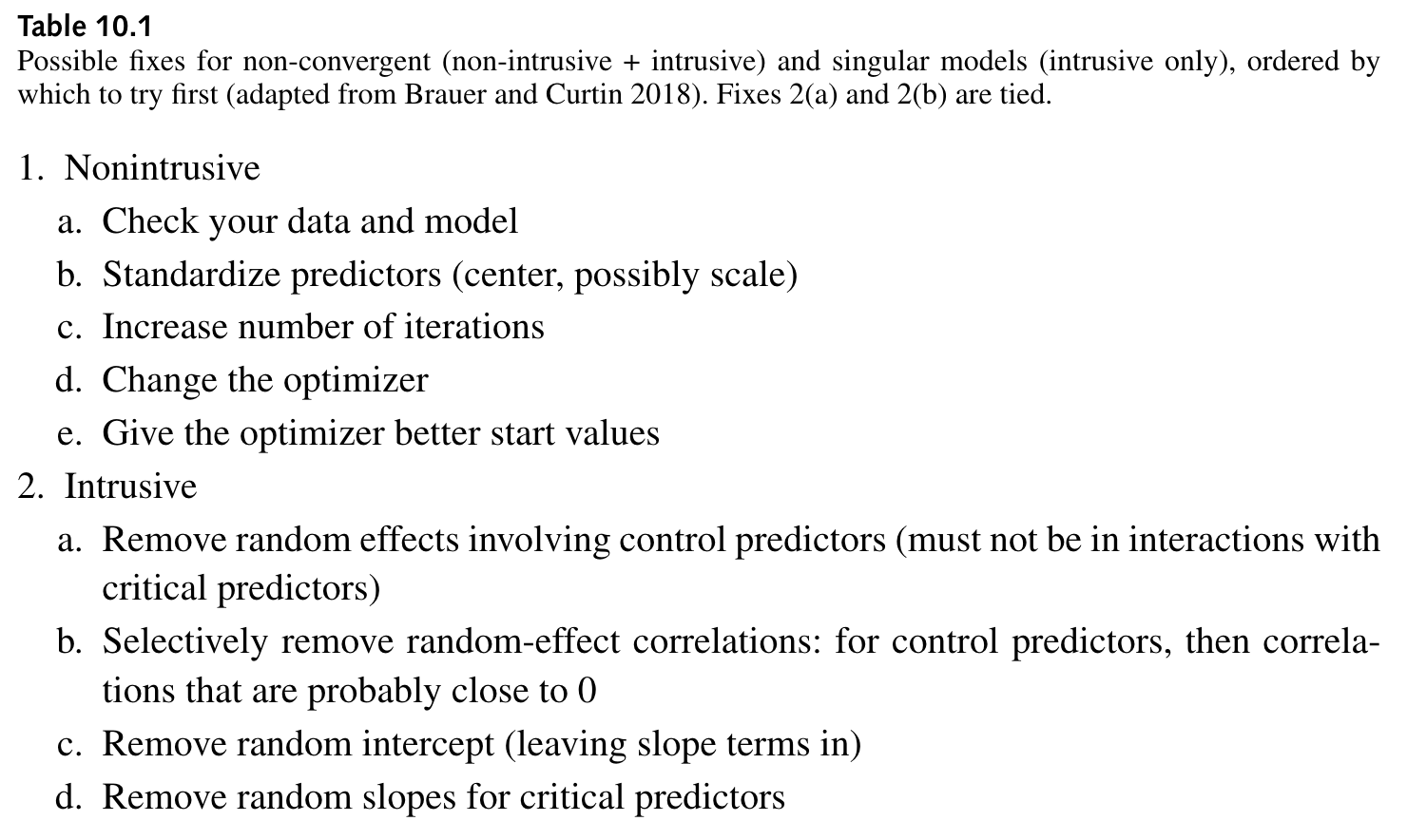
non-intrusive remedies amount to checking/adjusting data and model specifications
intrusive remedies involve reducing random effects structure
each strategy has its drawback
?convergence?convergence in the Console and read the vignette
?isSingularlme4::allFit(model) (can take a while to run)all_fit_verb_fp_mm <- allFit(fit_verb_fp_mm)
## bobyqa : boundary (singular) fit: see help('isSingular')
## [OK]
## Nelder_Mead : [OK]
## nlminbwrap : boundary (singular) fit: see help('isSingular')
## [OK]
## nmkbw : [OK]
## optimx.L-BFGS-B : boundary (singular) fit: see help('isSingular')
## [OK]
## nloptwrap.NLOPT_LN_NELDERMEAD : boundary (singular) fit: see help('isSingular')
## [OK]
## nloptwrap.NLOPT_LN_BOBYQA : boundary (singular) fit: see help('isSingular')
## [OK]
## There were 11 warnings (use warnings() to see them)default optimizer for lmer() is nloptwrap, formerly bobyqa (Bound Optimization by Quaradric Approximiation)
bobyqa helpssee ?lmerControl for more info
if fits are very similar (or all optimizeres except the default), the nonconvergent fit was a false positive
bobyqa Nelder_Mead
-2105.109 -2179.479
nlminbwrap nmkbw
-2105.106 -2105.109
optimx.L-BFGS-B nloptwrap.NLOPT_LN_NELDERMEAD
-2105.106 -2105.106
nloptwrap.NLOPT_LN_BOBYQA
-2105.106 (Intercept) verb_t1 gramm1 verb_t1:gramm1
bobyqa 5.956403 0.06170602 0.003369634 -0.01418865
Nelder_Mead 5.956350 0.06188102 0.003488675 -0.01397531
nlminbwrap 5.956403 0.06170726 0.003369637 -0.01419047
nmkbw 5.956404 0.06170653 0.003369153 -0.01419036
optimx.L-BFGS-B 5.956403 0.06170717 0.003369787 -0.01419044
nloptwrap.NLOPT_LN_NELDERMEAD 5.956403 0.06170725 0.003369649 -0.01419046
nloptwrap.NLOPT_LN_BOBYQA 5.956403 0.06170771 0.003369203 -0.014191841e5 in scientific notation)lmerControl()summary(rePCA(model)), lme4 package)
VarCorr(model))$item
Importance of components:
[,1] [,2] [,3] [,4]
Standard deviation 0.3638 0.2493 0.08366 0.000000000000000001309
Proportion of Variance 0.6567 0.3085 0.03474 0.000000000000000000000
Cumulative Proportion 0.6567 0.9653 1.00000 1.000000000000000000000
$sj
Importance of components:
[,1] [,2] [,3] [,4]
Standard deviation 0.6490 0.01470 0.000001281 0.00000001467
Proportion of Variance 0.9995 0.00051 0.000000000 0.00000000000
Cumulative Proportion 0.9995 1.00000 1.000000000 1.00000000000VarCorr(fit_verb_fp_mm) Groups Name Std.Dev. Corr
item (Intercept) 0.139189
verb_t1 0.055890 0.488
gramm1 0.022569 -0.109 -0.921
verb_t1:gramm1 0.095314 -0.283 0.456 -0.646
sj (Intercept) 0.257535
verb_t1 0.018296 0.974
gramm1 0.012055 0.960 0.872
verb_t1:gramm1 0.017731 0.991 0.934 0.990
Residual 0.399095 gramm because it has the lowest variance, and has a pretty high correlation with verb_t (which is unlikely to be true)gramm for participant for the same reason, as well as its high correlation with the intercept and verb_tfit_verb_fp_m1 <- lmer(log(fp) ~ verb_t*gramm +
(1 + verb_t+gramm|sj) +
(1 + verb_t+gramm|item),
data = df_biondo,
subset = roi == 4,
control = lmerControl(optimizer = "bobyqa",
optCtrl = list(maxfun = 2e5))
)boundary (singular) fit: see help('isSingular')rePCA()$item
Importance of components:
[,1] [,2] [,3]
Standard deviation 0.3559 0.1291 0.00000007181
Proportion of Variance 0.8837 0.1163 0.00000000000
Cumulative Proportion 0.8837 1.0000 1.00000000000
$sj
Importance of components:
[,1] [,2] [,3]
Standard deviation 0.6465 0.0000006824 0
Proportion of Variance 1.0000 0.0000000000 0
Cumulative Proportion 1.0000 1.0000000000 1VarCorr() Groups Name Std.Dev. Corr
item (Intercept) 0.139274
verb_t1 0.055550 0.489
gramm1 0.020747 -0.117 -0.924
sj (Intercept) 0.257657
verb_t1 0.017584 1.000
gramm1 0.011554 1.000 1.000
Residual 0.399869 fit_verb_fp_m2 <- lmer(log(fp) ~ verb_t*gramm +
(1 |sj) +
(1 + verb_t+gramm|item),
data = df_biondo,
subset = roi == 4,
control = lmerControl(optimizer = "bobyqa",
optCtrl = list(maxfun = 2e5))
)boundary (singular) fit: see help('isSingular')rePCA()$item
Importance of components:
[,1] [,2] [,3]
Standard deviation 0.3559 0.1297 0.00000001647
Proportion of Variance 0.8827 0.1173 0.00000000000
Cumulative Proportion 0.8827 1.0000 1.00000000000
$sj
Importance of components:
[,1]
Standard deviation 0.6441
Proportion of Variance 1.0000
Cumulative Proportion 1.0000VarCorr() Groups Name Std.Dev. Corr
item (Intercept) 0.139364
verb_t1 0.055805 0.485
gramm1 0.020546 -0.097 -0.917
sj (Intercept) 0.257648
Residual 0.399995 gramm and verb_t are highly correlatedgramm has least variance, so let’s remove itrePCA()$item
Importance of components:
[,1] [,2]
Standard deviation 0.3553 0.10311
Proportion of Variance 0.9223 0.07768
Cumulative Proportion 0.9223 1.00000
$sj
Importance of components:
[,1]
Standard deviation 0.6438
Proportion of Variance 1.0000
Cumulative Proportion 1.0000VarCorr()verb_t, so let’s run that modelfit_verb_fp_m4 <- lmer(log(fp) ~ verb_t*gramm +
(1 |sj) +
(1 + gramm|item),
data = df_biondo,
subset = roi == 4,
control = lmerControl(optimizer = "bobyqa",
optCtrl = list(maxfun = 2e5))
)boundary (singular) fit: see help('isSingular')verb_t slopessummary()Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: log(fp) ~ verb_t * gramm + (1 | sj) + (1 + verb_t | item)
Data: df_biondo
Control: lmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 200000))
Subset: roi == 4
REML criterion at convergence: 4216.2
Scaled residuals:
Min 1Q Median 3Q Max
-4.1758 -0.6096 -0.0227 0.6060 4.0568
Random effects:
Groups Name Variance Std.Dev. Corr
item (Intercept) 0.019423 0.13936
verb_t1 0.002513 0.05013 0.54
sj (Intercept) 0.066417 0.25771
Residual 0.160252 0.40032
Number of obs: 3795, groups: item, 96; sj, 60
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 5.956384 0.036763 79.243172 162.021 < 0.0000000000000002
verb_t1 0.061733 0.013971 93.410519 4.419 0.0000267
gramm1 0.003298 0.012999 3544.451690 0.254 0.80
verb_t1:gramm1 -0.014380 0.025998 3544.762213 -0.553 0.58
(Intercept) ***
verb_t1 ***
gramm1
verb_t1:gramm1
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) vrb_t1 gramm1
verb_t1 0.077
gramm1 0.000 -0.002
vrb_t1:grm1 0.000 0.002 0.000rePCA() + VarCorr() -> run model -> … -> converges -> only NOW run summary(model)sum_fit_verb_fp <-
tidy(fit_verb_fp,
effects = "fixed") |>
as_tibble() |>
mutate(p_value = p.value,
model = "parsimonious")
sum_fit_verb_fp_mm <-
tidy(fit_verb_fp_mm,
effects = "fixed") |>
as_tibble() |>
mutate(p_value = p.value,
model = "maximal")
sum_fit_verb_fp_intercepts <-
tidy(fit_verb_fp_intercepts,
effects = "fixed") |>
as_tibble() |>
mutate(p_value = p.value,
model = "intercepts")| term | parsimonious | intercepts | maximal | measure |
|---|---|---|---|---|
| (Intercept) | 5.9564 | 5.9564 | 5.9564 | estimate |
| verb_t1 | 0.0617 | 0.0619 | 0.0617 | estimate |
| gramm1 | 0.0033 | 0.0032 | 0.0034 | estimate |
| verb_t1:gramm1 | -0.0144 | -0.0143 | -0.0142 | estimate |
| term | parsimonious | intercepts | maximal | measure |
|---|---|---|---|---|
| (Intercept) | 0.0368 | 0.0368 | 0.0367 | std.error |
| verb_t1 | 0.0140 | 0.0130 | 0.0144 | std.error |
| gramm1 | 0.0130 | 0.0130 | 0.0133 | std.error |
| verb_t1:gramm1 | 0.0260 | 0.0260 | 0.0278 | std.error |
verb_t slopes:
| term | parsimonious | intercepts | maximal | measure |
|---|---|---|---|---|
| (Intercept) | 162.0213 | 161.9025 | 162.1605 | statistic |
| verb_t1 | 4.4188 | 4.7517 | 4.2982 | statistic |
| gramm1 | 0.2537 | 0.2466 | 0.2542 | statistic |
| verb_t1:gramm1 | -0.5531 | -0.5496 | -0.5108 | statistic |
verb_t: \(t_{max}\) < \(t_{pars}\) < \(t_{int}\)| term | parsimonious | intercepts | maximal | measure |
|---|---|---|---|---|
| (Intercept) | 79.2432 | 79.2008 | 79.1789 | df |
| verb_t1 | 93.4105 | 3637.1332 | 71.4491 | df |
| gramm1 | 3544.4517 | 3637.1834 | 179.9254 | df |
| verb_t1:gramm1 | 3544.7622 | 3637.1023 | 91.8597 | df |
lmerTest::lmer())
verb_t: \(df_{max}\) < \(df_{pars}\) < \(df_{int}\)
| term | parsimonious | intercepts | maximal | measure |
|---|---|---|---|---|
| (Intercept) | 0.0000000 | 0.0000000 | 0.0000000 | p.value |
| verb_t1 | 0.0000267 | 0.0000021 | 0.0000535 | p.value |
| gramm1 | 0.7997645 | 0.8052568 | 0.7996181 | p.value |
| verb_t1:gramm1 | 0.5802114 | 0.5826522 | 0.6107496 | p.value |
verb_t: \(p_{max}\) < \(p_{pars}\) < \(p_{int}\)
We included Time Reference (past, future), and Verb Match (match, mismatch) as fixed-effect factors in the models used to investigate the processing of past–future violations (Q1), by adopting sum contrast coding (Schad et al., 2020): past and match conditions were coded as –.5. while future and mismatch conditions were coded as .5. […] Moreover, we included crossed random intercepts and random slopes for all fixed-effect parameters for subject and item grouping factors (Barr et al., 2013) in all models.
We reduced the complexity of the random effect structure of the maximal model by performing a principal component analysis so as to identify the most parsimonious model properly supported by the data (Bates et al., 2015). […] all reading time data were log transformed before performing the analyses.
— Biondo et al. (2022), p. 9
We can create our own function, which we will call format_pval(), to produce formatted p-values. Here we use the case_when() function to print < .05, < .01 or < .001 when the p-value is smaller than these values (which is a convention). Otherwise (TRUE), round the p-value to 3 decimal points.
We can now use our format_pval() function to format our p-values and print them in a table, as in Table 13.6.
| effect | term | estimate | std.error | statistic | df | p_value |
|---|---|---|---|---|---|---|
| fixed | (Intercept) | 5.9563839 | 0.0367630 | 162.0213327 | 79.24317 | < .001 |
| fixed | verb_t1 | 0.0617330 | 0.0139706 | 4.4187865 | 93.41052 | < .001 |
| fixed | gramm1 | 0.0032976 | 0.0129994 | 0.2536709 | 3544.45169 | .8 |
| fixed | verb_t1:gramm1 | -0.0143804 | 0.0259984 | -0.5531269 | 3544.76221 | .58 |
The guidelines from the 7th edition from the American Psychological Association pertaining to numbers and statistics define how we should be formatting p-values, summarised in the Guide for Numbers and Statistics (American Psychological Association, 2022). A summary:
We used the round() function from base R to round p-values to 3 decimal points. This function drops trailing 0’s, so 0.030 will become 0.03. We also use the str_remove() function from the stringr package (Tidyverse), which takes the results from round() and removes any leading 0’s (i.e., those before the decimal) as defined by the regular expression (regex) ^0+. This will take 0.03 and print .03.
Here is some example output using round():
And removing the leading 0 using str_remove():
If our p-value is smaller than .001, then it will be written as < .001:
So now our p-values are formatted according to APA 7 guidelines, as long as we pass them through format_pval().
lme4brms R package)
Today we learned…
and how to…
| Term | Definition | Equation/Code |
|---|---|---|
| linear mixed (effects) model | NA | NA |
---
title: "Model selection"
subtitle: "Strolling through the garden of forking paths"
author: "Daniela Palleschi"
institute: Humboldt-Universität zu Berlin
# footer: "Lecture 1.1 - R und RStudio"
lang: en
date: "02/09/2024"
date-modified: last-modified
shift-heading-level-by: +1
---
::: {.callout-warning}
# Under construction {.unnumbered .uncounted .unlisted}
This chapter is not fully translated from bullet points (from my slides) to prose. This will happen eventually (hopefully by spring 2024).
:::
```{r setup, eval = T, echo = F}
knitr::opts_chunk$set(echo = T, # print chunks?
eval = T, # run chunks?
error = F, # print errors?
warning = F, # print warnings?
message = F, # print messages?
cache = F # cache?; be careful with this!
)
```
# Learning Objectives {.unnumbered .unlisted}
Today we will learn about...
- the history of mixed models (again)
- strategies for model selection
- variability in model selection
and how to...
- apply remedies for nonconvergence
- reduce our RES with a data-driven approach
- compare a parsimonious model to maximal and intercept-only models
# Resources {.unnumbered .unlisted}
- relevant papers for this topic
- @barr_random_maximal_2013
- @bates_parsimonious_2015
- @matuschek_balancing_2017
- @brauer_linear_2018
- @meteyard_best_2020
- practical resources covered in the practical application:
+ Sections 10.3-5 in @sonderegger_regression_2023
+ Section 15.7.3 'Convergence Issues' in @winter_statistics_2019
+ @brauer_linear_2018
+ @meteyard_best_2020
- we will continue using the data from @biondo_yesterday_2022
```{r hidden_packages}
#| echo: false
#| eval: true
# extra packages for the lecture notes/slides
pacman::p_load(
tidyverse,
here, janitor,
googlesheets4,
gt)
# tell googlesheets4 we don't want private
gs4_deauth()
```
```{r}
#| echo: false
#| eval: true
df_biondo <-
read_csv(here("data", "Biondo.Soilemezidi.Mancini_dataset_ET.csv"),
locale = locale(encoding = "Latin1") ## for special characters in Spanish
) |>
clean_names() |>
mutate(gramm = ifelse(gramm == "0", "ungramm", "gramm")) |>
mutate_if(is.character,as_factor) |> # all character variables as factors
droplevels() |>
filter(adv_type == "Deic")
```
# Review: random intercepts and slopes
- violation of independence assumption --> increased Type I error (false positive)
+ include random effects per plausible grouping factor [herein "unit", à la @barr_random_maximal_2013]
- random-intercepts only models --> increased Type I error (false positive)
+ add random slopes per within-unit manipulation, i.e., "maximal" models [@barr_random_maximal_2013]
- but such models often fail to "converge"
+ and have been shown to increase Type II error (false negative) [@bates_parsimonious_2015; @matuschek_balancing_2017]
- some questions remain:
+ how to define our maximal model
+ how to handle convergence issues
# History of LMMs revisited
- recall @clark_language-as-fixed-effect_1973's language-as-fixed-effect fallacy and the issue of generalisability [see also @winter_independence_2021; @yarkoni_generalizability_2022]
- @baayen_mixed-effects_2008 motivated LMM's for linguistic data in the JML special issue
+ effect: everybody adopted random-intercepts only models
- @barr_random_maximal_2013: random-intercepts only models are overconfident, "keep it maximal!"
+ effect: everybody adopted maximal models
- @matuschek_balancing_2017 and @bates_parsimonious_2015: maximal models are underconfident and lower statistical power! Use data-driven model selection to find a "parsimonious" model!
+ effect: some people adopt this method, but many psycholinguists just want a "recipe" to follow
## 2013: Keep it maximal
> A maximal model should optimize generalization of the findings to new subjects and new items.
>
> -- @barr_random_maximal_2013, p. 261
- random-intercepts-only models tend to be underpowered
- for this reason, @barr_random_maximal_2013 suggested using a maximal random effects structure justified by the experimental design
## 2015 & 2017: Parsimonious models
> [W]hile the maximal model indeed performs well as far as Type I error rates were concerned, power decreases substantially with model complexity.
>
> --- @matuschek_balancing_2017, p. 310-311
- there is a trade-off between Type I (overconfidence) and Type II error (underconfidence)
- i.e., maximal models can lead to over-fitting
+ lowers statistical power, which increases Type II error (false rejection)
- but we should strive for the most *parsimonious* model
+ parsimonous models are a compromise between the maximal model justified by your design and theory, and a given data set
- best way to maintain low Type I and II error: collect lots of data
# Model building
> Every statistical model is a description of some real or hypothetical state of affairs in the world.
>
> -- @yarkoni_generalizability_2022, p. 2
- our models reflect not only our hypotheses or effects of interest, but any other plausible or known co-variates
- our predictors should reflect our research questions or theories tested
+ plus any plausible or previously motivated co-variates (e.g., trial order)
+ plus known sources of nonindependence, i.e., our random effects
## Choosing predictors
- your model should be defined *a priori*
+ i.e., you should define what predictors you will include and any covariates
+ e.g., if you have a prediction about the effect of phonological neighbours on vowel duration
+ define what phonological characteristics you will include (e.g., place of articulation? manner?)
+ these should be related to specific hypotheses/research questions
## Choosing a maximal random effects structure (RES)
- how do we define our maximal model? Some tips from @barr_random_maximal_2013
+ between-unit factor (e.g., age): include random intercept only
+ within-unit factor with multiple observations per unit-level (e.g., age in longitudinal data): include random slopes
- all factors in an interaction are within-unit: include by-unit random slopes for interaction terms
### Example: @biondo_yesterday_2022
- @biondo_yesterday_2022: 2x2 design
+ verb-tense (past, future) and grammaticality (grammatical, ungrammatical)
+ repeated-measures: within-participant and -item design
+ so we should have by-participant and -item random intercepts (multiple observations per unit level)
+ each participant and item contributed multiple data points *per condition* (i.e., tense and grammaticality were manipulated within each unit level)
+ so we should have varying tense and grammaticality slopes by- item and -participant
```{r}
#| eval: false
#| warning: true
#| output-location: fragment
#| code-fold: true
fit_verb_fp_mm <- lmer(log(fp) ~ verb_t*gramm +
(1 + verb_t*gramm|sj) +
(1 + verb_t*gramm|item),
data = df_biondo,
subset = roi == 4)
```
## Observations per cell
- if there is only a single observation per cell, e.g., you collected one observation from each participant per condition, then you can't fit random intercepts or slopes
- ideally you would have at least 5 observations per cell (per unit level per condition, e.g., each participant has at least 5 observations per condition)
- this is also a question of statistical power
```{r}
#| output-location: column-fragment
#| eval: true
# obvz per sj per condition
df_biondo |>
filter(roi == 4) |>
count(sj, verb_t, gramm) |>
count(n)
```
```{r}
#| eval: true
#| output-location: column-fragment
# obvz per item per condition
df_biondo |>
filter(roi == 4) |>
count(item, verb_t, gramm) |>
arrange(desc(n)) |>
count(n)
```
## Data structure
- random effects must be factors/categorical
- single observation per row
+ generally speaking, there should be n(participants) * n(items) rows
+ every fixed or random effect in your model should correspond to a column in your dataset
# Variability in methods
- @meteyard_best_2020
+ survey of (psychology) researchers
+ review of papers using LMMs
- insecurity in researchers re: choosing models
- great variation in papers in how models are built and reported
## Researcher degrees of freedom
> What we hope to make clear is that there is no single correct way in which LMM analyses should be conducted, and this has important implications for how the reporting of LMMs should be approached.
>
> --- @meteyard_best_2020, p. 9
- the problem:
+ 'researcher degrees of freedom' [@simmons_false-positive_2011], or 'the garden of forking paths' [@gelman_garden_2013]
+ the same data can be analysed in a variety of ways
- this leads to insecurity for many researchers
## Justify and document
> Replicability and reproducibility are critical for scientific progress, so the way in which researchers have implemented LMM analysis must be entirely transparent. We also hope that the sharing of analysis code and data becomes widespread, enabling the periodic re-analysis of raw data over multiple experiments as studies accumulate over time.
>
> --- @meteyard_best_2020, p. 9
- the (partial) solution:
+ make model building/selection decisions a priori
+ be transparent
+ share your data and code
# A practical example
We'll now look at an example of a model that encounters convergence issues, and take some steps to reach convergence. We are considering a case where your predictors and covariates are already selected, and will focus on selecting model options and random effects structure that achieve model convergence.
## Set-up
We first need to set up our environment.
```{r}
## suppress scientific notation
options(scipen=999)
```
### Load packages {.unnumbered}
```{r}
#| eval: true
## load libraries
pacman::p_load(
tidyverse,
here,
janitor,
## new packages for mixed models:
lme4,
lmerTest,
broom.mixed,
lattice)
```
```{r}
#| echo: false
## set preferred ggplot2 theme
theme_set(theme_bw() + theme(plot.title = element_text(size = 10)))
```
```{r}
lmer <- lmerTest::lmer
```
```{r}
#| echo: false
## extra packages for the lecture notes/slides
pacman::p_load(
patchwork,
knitr,
kableExtra,
googlesheets4,
gt)
## tell googlesheets4 we don't want private
gs4_deauth()
```
### Load data {.unnumbered}
- data from @biondo_yesterday_2022
```{r}
df_biondo <-
read_csv(here("data", "Biondo.Soilemezidi.Mancini_dataset_ET.csv"),
locale = locale(encoding = "Latin1") ### for special characters in Spanish
) |>
clean_names() |>
mutate(gramm = ifelse(gramm == "0", "ungramm", "gramm")) |>
mutate_if(is.character,as_factor) |> ## all character variables as factors
droplevels() |>
filter(adv_type == "Deic")
```
### Set contrasts
```{r}
contrasts(df_biondo$verb_t) <- c(-0.5,+0.5)
contrasts(df_biondo$gramm) <- c(-0.5,+0.5)
```
```{r}
contrasts(df_biondo$verb_t)
```
```{r}
contrasts(df_biondo$gramm)
```
## Start maximal
- model structure should be decided *a priori*
+ included fixed (predictors and covariates) and random effects
### Maximal model
- starting point: most maximal model structure justified by your design
+ if this converges, great!
+ if it doesn't, what does this mean and what should we do?
```{r}
#| eval: false
#| warning: true
#| message: true
#| output-location: fragment
fit_verb_fp_mm <- lmer(log(fp) ~ verb_t*gramm +
(1 + verb_t*gramm|sj) +
(1 + verb_t*gramm|item),
data = df_biondo,
subset = roi == 4)
```
- we get a warning of singular fit
```{r}
#| echo: false
#| eval: true
## saveRDS(fit_verb_fp_mm, here::here("slides", "12-model_selection_example", "rds", "fit_verb_fp_mm.rds"))
fit_verb_fp_mm <- readRDS(here::here("slides", "12-model_selection_example", "rds", "fit_verb_fp_mm.rds"))
```
## Convergence issues
- "*Convergence is not a metric of model quality*" [@sonderegger_regression_2023, p. 365, Box 10.2]
+ convergence does not always indicate "overfitting" or "overparameterisation"
+ can also be due to optimizer choice
+ since default optimizer was changed to `nloptwrap` from `bobyqa`, there seem to be more 'false positive' convergence warnings
- false-positive convergence: you get a convergence warning, but changing the optimizer and/or iteration count does not produce a warning
- false-negative convergence: you do not get a warning, but your variance-covariance matrix might indicate overfitting
### Nonconvergence remedies
- unfortunately there is no one "right" way to deal with convergence issues
+ important is to transparently report and justify your method
- Table 17 in @brauer_linear_2018 (p. 404) suggests 20 remedies, whittled down to 10 suggestions in @sonderegger_regression_2023
```{r}
#| label: fig-table10
#| fig-cap: "From @sonderegger_regression_2023, p. 366"
#| echo: false
include_graphics(here::here("media", "sonderegger_2023_convergence_table10.png"))
```
### Intrusive vs. Non-intrusive remedies
- non-intrusive remedies amount to checking/adjusting data and model specifications
- intrusive remedies involve reducing random effects structure
+ there are different schools of thought
+ random-intercepts only: increased Type I error rate = overconfident estimates
+ maximal-but-singular-fit model [@barr_random_maximal_2013]: reduces power = underconfident estimates
+ data-driven approach [@bates_parsimonious_2015]: can lose the forest for the trees, e.g., removing random slopes for predictors of interest
- each strategy has its drawback
+ important is to choose your strategy *a priori* and transparently report and justify your strategy
+ even better: share/publish your data and code, which should be reproducible
### `?convergence`
- type `?convergence` in the Console and read the vignette
+ what suggestions does it make?
- compare this to `?isSingular`
## Non-intrusive methods
- check your data structure/variables
+ check model assumptions (e.g., normality, missing transformations of variables)
+ check your RES is justified by your experimental design/data structure
+ centre your predictors (e.g., sum contrasts, or centring/standardizing) to reduce multicollinearity; reduces collinearity in the random effects (a possible source of nonconvergence)
+ check observations per cell (e.g., is there a participant very few observations, or few observations per one condition? Should be at least >5 per cell)
- alter model controls:
- increase iterations
- check optimizer
### Check optimzer
- optimizer
+ `lme4::allFit(model)` (can take a while to run)
```{r}
#| output-location: column-fragment
#| eval: false
all_fit_verb_fp_mm <- allFit(fit_verb_fp_mm)
## bobyqa : boundary (singular) fit: see help('isSingular')
## [OK]
## Nelder_Mead : [OK]
## nlminbwrap : boundary (singular) fit: see help('isSingular')
## [OK]
## nmkbw : [OK]
## optimx.L-BFGS-B : boundary (singular) fit: see help('isSingular')
## [OK]
## nloptwrap.NLOPT_LN_NELDERMEAD : boundary (singular) fit: see help('isSingular')
## [OK]
## nloptwrap.NLOPT_LN_BOBYQA : boundary (singular) fit: see help('isSingular')
## [OK]
## There were 11 warnings (use warnings() to see them)
```
```{r}
#| echo: false
## saveRDS(all_fit_verb_fp_mm, here::here("slides", "12-model_selection_example", "rds", "all_fit_verb_fp_mm.rds"))
all_fit_verb_fp_mm <- readRDS(here::here("slides", "12-model_selection_example", "rds", "all_fit_verb_fp_mm.rds"))
```
### Optimizers
+ default optimizer for `lmer()` is `nloptwrap`, formerly `bobyqa` (Bound Optimization by Quaradric Approximiation)
+ usually changing to `bobyqa` helps
+ see `?lmerControl` for more info
- if fits are very similar (or all optimizeres except the default), the nonconvergent fit was a false positive
+ it's safe to use the new optimizer
```{r}
summary(all_fit_verb_fp_mm)$llik
```
```{r}
summary(all_fit_verb_fp_mm)$fixef
```
### Increase iterations
- and/or increase number of iterations
+ default is 10 000 (`1e5` in scientific notation)
+ you can try 20 000, 100 000, etc.
+ this usually helps with larger data or models with complex RES
```{r}
## check n of iterations
fit_verb_fp_mm@optinfo$feval
```
### `lmerControl()`
```{r}
#| eval: false
#| warning: true
#| output-location: fragment
fit_verb_fp_mm <- lmer(log(fp) ~ verb_t*gramm +
(1 + verb_t*gramm|sj) +
(1 + verb_t*gramm|item),
data = df_biondo,
subset = roi == 4,
control = lmerControl(optimizer = "bobyqa",
optCtrl = list(maxfun = 2e5))
)
```
- or you can just 'update' the model to save some syntax
```{r}
#| eval: true
#| warning: true
#| output-location: fragment
fit_verb_fp_mm <- update(fit_verb_fp_mm,
control = lmerControl(optimizer = "bobyqa",
optCtrl = list(maxfun = 2e5)))
```
### Removing parameters
- still won't converge?
+ it's time to consider intrusive remedies: removing random effects parameters
## Intrusive methods
- nonconvergence in maximal models is often due to overfitting
+ i.e., the model is overly complex given your data
+ this is typically due to an overly complex random effects structure
- if the non-intrusive methods don't lead to convergence, the problem is likely overfitting
### Parsimonious vs. maximal
- there are different camps on how to deal with this issue
- I personally follow the suggestions in @bates_parsimonious_2015 (for now)
1. run random effects Principal Components Analysis (`summary(rePCA(model))`, `lme4` package)
+ informs by how many parameters our model is overfit
2. check variance-covariance matrix (`VarCorr(model)`)
3. remove parameters with very high or low Correlation terms and/or much lower variance compared to other terms
4. fit simplified model
5. wash, rinse, repeat
- we'll practice this method today, but keep in mind that it's up to you to decide and justify which method you use
### Random effects Principal Components Analysis
- gives us a ranking of all parameters ('components') in our RES per unit
```{r}
#| output-location: fragment
summary(rePCA(fit_verb_fp_mm))
```
###
- important is the Cumulative Proportion
+ how much of the cumulative variance explained by all the by-unit parameters does this one parameter contribute?
+ we see for item, the first component accounts for 66% of the variance explained, and the next contributes an additional 31%, and the next 3%
+ so two components account for roughly 97% of variance explained by our RES
+ in other words, we can remove one component for sure, and possibly another
+ we could potentially remove 3 components from participant
### Variance-covariance matrix
- so we can remove 2 parameters from item and participant
+ so either the varying intercept, or slope for tense, grammaticality, or their interaction
- we can check this with `VarCorr(fit_verb_fp_mm)`
```{r}
#| output-location: fragment
VarCorr(fit_verb_fp_mm)
```
- for item I would remove `gramm` because it has the lowest variance, and has a pretty high correlation with `verb_t` (which is unlikely to be true)
- I would also remove `gramm` for participant for the same reason, as well as its high correlation with the intercept and `verb_t`
#### Alternate model 1
- for now let's just remove the interaction term
+ for reproducibility reasons, do not delete the code for a model that did not converge
+ rather, write a comment on what decision was made (and why) for the new model
```{r}
#| message: true
#| warning: true
#| output-location: fragment
fit_verb_fp_m1 <- lmer(log(fp) ~ verb_t*gramm +
(1 + verb_t+gramm|sj) +
(1 + verb_t+gramm|item),
data = df_biondo,
subset = roi == 4,
control = lmerControl(optimizer = "bobyqa",
optCtrl = list(maxfun = 2e5))
)
```
##### `rePCA()`
```{r}
#| output-location: fragment
summary(rePCA(fit_verb_fp_m1))
```
##### `VarCorr()`
```{r}
#| output-location: fragment
VarCorr(fit_verb_fp_m1)
```
- when we see Corr +/-1, this tells us there was an error computing correlations between parameters
+ it is an invitation to explore
- this is not plausible, and indicates overfitting in our model
+ we can remove all slopes from sj
##### by-item random effects
```{r}
lattice::dotplot(ranef(fit_verb_fp_m1))$item
```
##### by-participant random effects (with +1 correlations)
```{r}
lattice::dotplot(ranef(fit_verb_fp_m1))$sj
```
#### Alternate model 2
```{r}
#| message: true
#| warning: true
#| output-location: fragment
fit_verb_fp_m2 <- lmer(log(fp) ~ verb_t*gramm +
(1 |sj) +
(1 + verb_t+gramm|item),
data = df_biondo,
subset = roi == 4,
control = lmerControl(optimizer = "bobyqa",
optCtrl = list(maxfun = 2e5))
)
```
##### `rePCA()`
```{r}
summary(rePCA(fit_verb_fp_m2))
```
##### `VarCorr()`
```{r}
VarCorr(fit_verb_fp_m2)
```
- by-item slopes for `gramm` and `verb_t` are highly correlated
- `gramm` has least variance, so let's remove it
##### by-item random effects
```{r}
lattice::dotplot(ranef(fit_verb_fp_m2))$item
```
#### Alternate model 3
```{r}
#| message: true
#| warning: true
#| output-location: fragment
fit_verb_fp_m3 <- lmer(log(fp) ~ verb_t*gramm +
(1 |sj) +
(1 + verb_t|item),
data = df_biondo,
subset = roi == 4,
control = lmerControl(optimizer = "bobyqa",
optCtrl = list(maxfun = 2e5))
)
```
- converged!
##### `rePCA()`
```{r}
summary(rePCA(fit_verb_fp_m3))
```
##### `VarCorr()`
```{r}
VarCorr(fit_verb_fp_m3)
```
#### Alternate model 4
- but we might've also decided to remove `verb_t`, so let's run that model
```{r}
#| message: true
#| warning: true
#| output-location: fragment
fit_verb_fp_m4 <- lmer(log(fp) ~ verb_t*gramm +
(1 |sj) +
(1 + gramm|item),
data = df_biondo,
subset = roi == 4,
control = lmerControl(optimizer = "bobyqa",
optCtrl = list(maxfun = 2e5))
)
```
- does not converge, so we're justified in keeping by-item `verb_t` slopes
### Final model
- the final model name should be some sort of convention to make your life easier
+ so remove model index
```{r}
fit_verb_fp <- fit_verb_fp_m3
```
##### by-item random effects
```{r}
lattice::dotplot(ranef(fit_verb_fp))$sj
```
##### by-participant random effects
```{r}
lattice::dotplot(ranef(fit_verb_fp))$item
```
##### `summary()`
```{r}
summary(fit_verb_fp)
```
- IMPORTANTLY, only look at the fixed effects after you've got your final model!!!!
+ i.e., run model -> convergence error -> `rePCA()` + `VarCorr()` -> run model -> ... -> converges -> only NOW run `summary(model)`
## Comparing to 'bad' models
- let's compare our final model to our 'bad' models
+ random intercepts-only model (overconfident)
+ maximal model (underconfident)
### Random-intercepts only
```{r}
#| message: true
#| warning: true
#| output-location: fragment
fit_verb_fp_intercepts <- lmer(log(fp) ~ verb_t*gramm +
(1 |sj) +
(1 |item),
data = df_biondo,
subset = roi == 4
)
```
- converges
```{r}
#| code-fold: true
sum_fit_verb_fp <-
tidy(fit_verb_fp,
effects = "fixed") |>
as_tibble() |>
mutate(p_value = p.value,
model = "parsimonious")
sum_fit_verb_fp_mm <-
tidy(fit_verb_fp_mm,
effects = "fixed") |>
as_tibble() |>
mutate(p_value = p.value,
model = "maximal")
sum_fit_verb_fp_intercepts <-
tidy(fit_verb_fp_intercepts,
effects = "fixed") |>
as_tibble() |>
mutate(p_value = p.value,
model = "intercepts")
```
### coefficient estimates {.smaller}
```{r}
#| label: tbl-estimates
#| code-fold: true
#| tbl-cap: "Coefficient estimates for our parsimonious model, a random-intercepts only model, and a maximal model"
rbind(sum_fit_verb_fp, sum_fit_verb_fp_intercepts, sum_fit_verb_fp_mm) |>
select(term, estimate, model) |>
mutate(estimate = round(estimate,4)) |>
pivot_wider(
id_cols = c(term),
names_from = model,
values_from = estimate
) |>
mutate(measure = "estimate") |>
kable() |>
kable_styling()
```
### standard error {.smaller}
```{r}
#| label: tbl-std_error
#| code-fold: true
#| tbl-cap: "Standard error of coefficient estimates for our parsimonious model, a random-intercepts only model, and a maximal model"
rbind(sum_fit_verb_fp, sum_fit_verb_fp_intercepts, sum_fit_verb_fp_mm) |>
select(term, std.error, model) |>
mutate(std.error = round(std.error,4)) |>
pivot_wider(
id_cols = c(term),
names_from = model,
values_from = std.error
) |>
mutate(measure = "std.error") |>
kable() |>
kable_styling()
```
- standard error (\\$SE = \frac{\sigma}{\sqrt{n}}\\$) is a measure of uncertainty
+ larger values reflect greater uncertainty
+ because $n$ is in the denominator, SE gets smaller with more observations
- compared to our parsimonious model with by-item varying `verb_t` slopes:
+ smaller SE for our overconfident (intercepts) model
+ larger SE for our underconfident (maximal) model
+ but only for the estimate also included in the random effects
### t-values {.smaller}
```{r}
#| label: tbl-t_value
#| code-fold: true
#| tbl-cap: "t-values of each estimates for our parsimonious model, a random-intercepts only model, and a maximal model"
rbind(sum_fit_verb_fp, sum_fit_verb_fp_intercepts, sum_fit_verb_fp_mm) |>
select(term, statistic, model) |>
mutate(statistic = round(statistic,4)) |>
pivot_wider(
id_cols = c(term),
names_from = model,
values_from = statistic
) |>
mutate(measure = "statistic") |>
kable() |>
kable_styling()
```
- t-value (\\$t = \frac{\bar{x}_1 - \bar{x}_2}{SE}\\$) is a measure of uncertainty
+ larger values reflect greater effect
+ more $n$ increases $t$
- again, `verb_t`: $t_{max}$ < $t_{pars}$ < $t_{int}$
### degrees of freedom {.smaller}
```{r}
#| label: tbl-df
#| code-fold: true
#| tbl-cap: "Degrees of freedom of each estimates for our parsimonious model, a random-intercepts only model, and a maximal model"
rbind(sum_fit_verb_fp, sum_fit_verb_fp_intercepts, sum_fit_verb_fp_mm) |>
select(term, df, model) |>
mutate(df = round(df,4)) |>
pivot_wider(
id_cols = c(term),
names_from = model,
values_from = df
) |>
mutate(measure = "df") |>
kable() |>
kable_styling()
```
- degrees of freedom: not trivially defined in mixed models; we're using Satterthwaite approximiation (default in `lmerTest::lmer()`)
+ larger degrees of freedom corresponds to larger $n$
+ including more random effects reduces our $n$ and therefore reduces $df$
- again, `verb_t`: $df_{max}$ < $df_{pars}$ < $df_{int}$
+ and large differences between our maximal model and the other two for other terms
### p-values {.smaller}
```{r}
#| label: tbl-p_value
#| code-fold: true
#| tbl-cap: "p-values of coefficient estimates for our parsimonious model, a random-intercepts only model, and a maximal model"
rbind(sum_fit_verb_fp, sum_fit_verb_fp_intercepts, sum_fit_verb_fp_mm) |>
select(term, p.value, model) |>
mutate(p.value = round(p.value, 8)) |>
pivot_wider(
id_cols = c(term),
names_from = model,
values_from = p.value
) |>
mutate(measure = "p.value") |>
kable() |>
kable_styling()
```
- p-values: inversely related to t-values (larger t-values = smaller p-values)
- again, `verb_t`: $p_{max}$ < $p_{pars}$ < $p_{int}$
+ this would be important for 'signicance' if the values were closer to the convential alpha-levels (p < .05, p < .01, p < .001)
+ but here the different random effects structures don't qualitatively change (all are < .001)
- this is not always the case, however!
+ this is why we do not peek at the fixed effects until we have our final model
+ we don't want to be influenced (consciously or not) by seeing small p-values in one model but not another
## Reporting
- in Data Analysis section, e.g.,
> We included Time Reference (past, future), and Verb Match (match, mismatch) as fixed-effect factors in the models used to investigate the processing of past–future violations (Q1), by adopting sum contrast coding (Schad et al., 2020): past and match conditions were coded as –.5. while future and mismatch conditions were coded as .5. [...] Moreover, we included crossed random intercepts and random slopes for all fixed-effect parameters for subject and item grouping factors (Barr et al., 2013) in all models.
>
> We reduced the complexity of the random effect structure of the maximal model by performing a principal component analysis so as to identify the most parsimonious model properly supported by the data (Bates et al., 2015). [...] all reading time data were log transformed before performing the analyses.
>
> --- @biondo_yesterday_2022, p. 9
### Formatted p-values
We can create our own function, which we will call `format_pval()`, to produce formatted p-values. Here we use the `case_when()` function to print `< .05`, `< .01` or `< .001` when the p-value is smaller than these values (which is a convention). Otherwise (`TRUE`), round the p-value to 3 decimal points.
```{r}
## function to format p-values to APA7 guidelines
format_pval <- function(pval){
dplyr::case_when(
pval < .001 ~ "< .001",
TRUE ~ str_remove(round(pval, 3), "^0+")
)
}
```
We can now use our `format_pval()` function to format our p-values and print them in a table, as in @tbl-format_pval.
```{r}
#| label: tbl-format_pval
#| tbl-cap: "Table with formatted p-values from `format_pval()`"
#| code-fold: true
#| eval: true
tidy(fit_verb_fp,
effects = "fixed") |>
as_tibble() |>
mutate(p_value = format_pval(p.value)) |>
select(-p.value) |>
knitr::kable() |>
kableExtra::kable_styling()
```
::: {.callout-tip}
# APA 7 guidelines: formatting p-values
The guidelines from the 7th edition from the American Psychological Association pertaining to numbers and statistics define how we should be formatting *p*-values, summarised in the [Guide for Numbers and Statistics](https://apastyle.apa.org/instructional-aids/numbers-statistics-guide.pdf) [@apa_numbers_2022]. A summary:
- Don't use leading 0's (i.e., zero before a decimal) when the statistic cannot be greater than 1 (proportion, correlation, level of statistical significance).
- Report exact *p*-values to two or three decimals (e.g., p = .006, p = .03). i.e., no trailing 0's
- Report *p*-values less than .001 as "p < .001".
- Do not repeat statistics in both the text and a table or figure
We used the `round()` function from base R to round *p*-values to 3 decimal points. This function drops trailing 0's, so `0.030` will become `0.03`. We also use the `str_remove()` function from the `stringr` package (Tidyverse), which takes the results from `round()` and removes any leading 0's (i.e., those before the decimal) as defined by the regular expression (`regex`) `^0+`. This will take `0.03` and print `.03`.
Here is some example output using `round()`:
```{r}
round(0.020001,3)
```
And removing the leading 0 using `str_remove()`:
```{r}
str_remove(round(0.020001,3), "^0+")
```
If our *p*-value is smaller than .001, then it will be written as `< .001`:
```{r}
format_pval(c(0.2, 0.02, 0.002, 0.0002))
```
So now our *p*-values are formatted according to APA 7 guidelines, as long as we pass them through `format_pval()`.
:::
# Alternatives to `lme4`
- other alternatives that have fewer convergence issues:
+ Julia
+ e.g., in VS Code IDE
+ Bayesian framework (e.g., `brms` R package)
+ also run (G)LMMs, but abandons arbitrary p-values
+ instead quantifies uncertainty
- both are more more computationally powerful
+ the are not (yet) as widely used in the field
# Learning objectives 🏁 {.unnumbered .unlisted .uncounted}
Today we learned...
- the history of mixed models (again) ✅
- strategies for model selection ✅
- variability in model selection ✅
and how to...
- apply remedies for nonconvergence ✅
- reduce our RES with a data-driven approach ✅
- compare a parsimonious model to maximal and intercept-only models ✅
# Important terms {.unnumbered .smaller .uncounted}
```{r terms}
#| echo: false
content <-
googlesheets4::read_sheet("https://docs.google.com/spreadsheets/d/17CqdxKL9lyy-PbTB2ZnfWNWs4oV--CcBvrqlh_aEPGQ/edit?usp=sharing")
content |>
filter(`Lecture topic` == "08 - LMMs 1: random intercepts") |>
select(-`Lecture topic`) |>
gt()
```
# References {.unlisted .unnumbered visibility="uncounted"}
::: {#refs custom-style="Bibliography"}
:::