Reproducibility in linguistic research and R
What, Why, and How?
Learning Objectives
Today we will learn about…
- what ‘reproducibility’ is and why/how to practice it
- FAIR principles for sharing our code and data
- concepts for building a reproducible workflow
1 What is Open Science?
“Open science” is an umbrella term used to refer to the concepts of openness, transparency, rigor, reproducibility, replicability, and accumulation of knowledge, which are considered fundamental features of science”
— Crüwell et al. (2019), p.3
- a movement developed to respond to crisis in scientific research
- lack of accessibility, transparency, reproducibility, and replicability of previous research
- transparency is key to all facets of Open Science
- it allows for full evaluation of all stages of science
- Open Access, software, data, code, materials…
1.1 Systemic problem in science
- the combination of
- publication bias
- journals favour novel, significant findings
- publish or perish
- researchers’ careers depend on publications
- publication bias
- can/does/did lead to:
- HARKing
- Hypothesising After Results are Known
- p-hacking
- (re-)running analyses until a significant effect is found
- replication crisis
- pervasive failure to replicate previous research
- HARKing
1.2 Why do Open Science?
- open science is good science
- it encourages organisation and planning
- helpful for future you
- increases transparency
- without transparency we cannot inspect evidence ourselves
- or ensure the claims match the evidence
- makes our work more robust
- so future work stands on solid ground
- not all-or-nothing
- there are things I consider the bare minimum
- detailed experiment plan, ideally public
- openly available materials (e.g., stimuli)
- share code and data
- the important thing is to do what you can
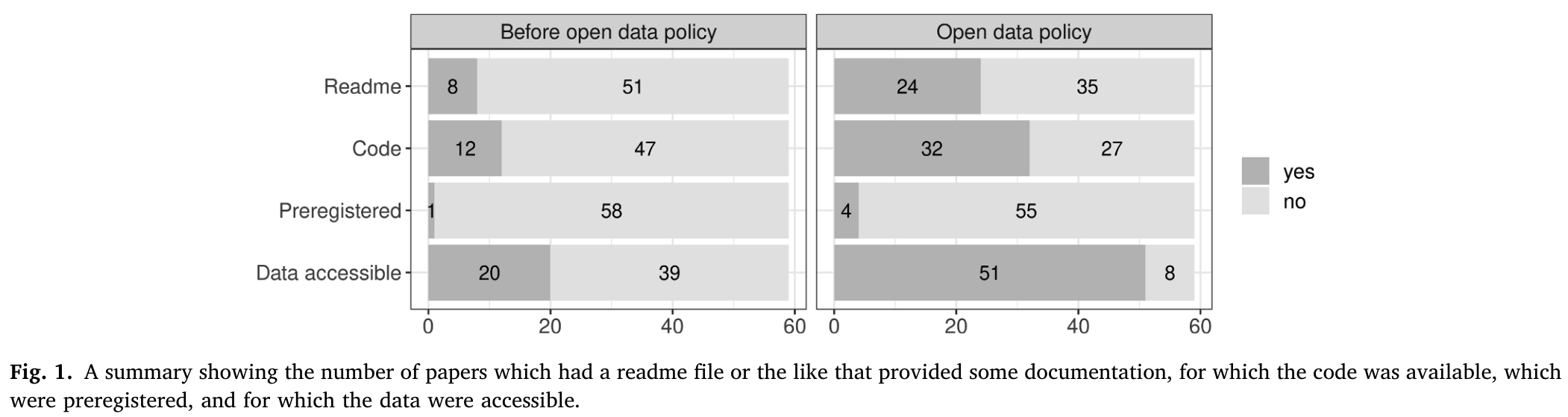
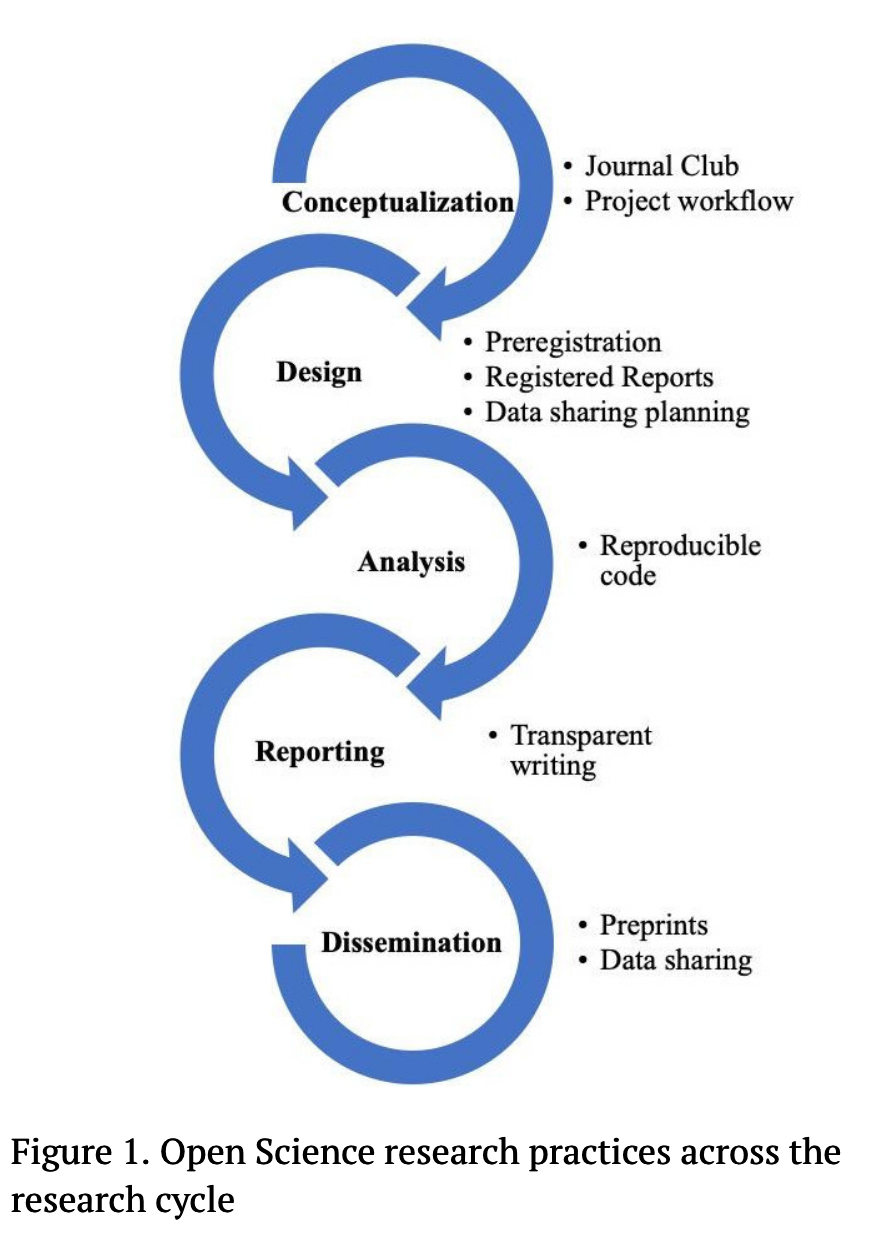
- which Open Science research practices in Figure 1 are relevant related to reproducibility?

The replication crisis refers to the failure of many replication studies to replicate the findings of influential studies. The result of this “crisis” is a move towards Open Science Practices, which emphasise transparency along all stages of research (conception, planning, data collection, data cleaning, data analysis, reporting).
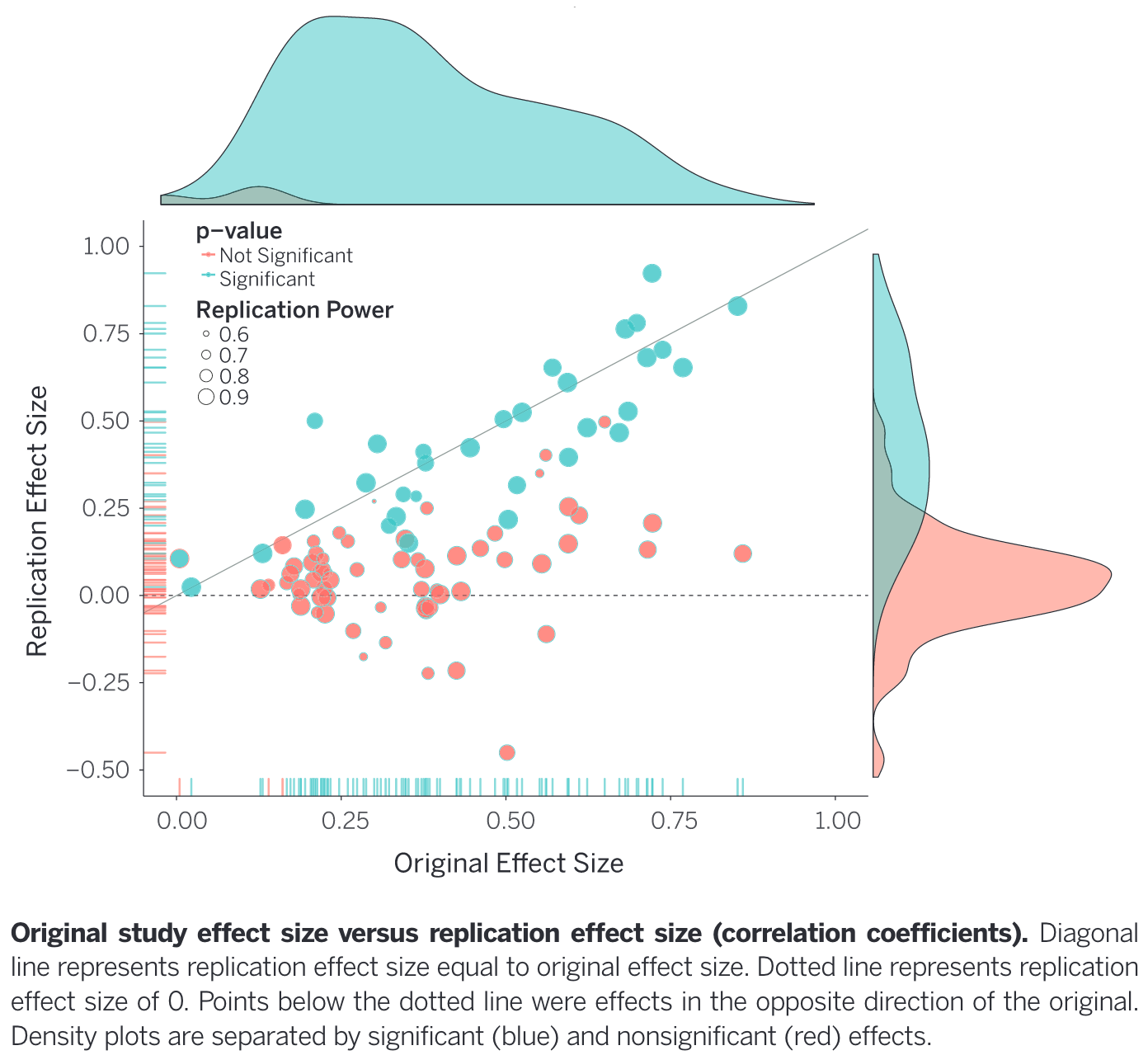
The issue became more widespread with the publication of Ioannidis (2005), entitled Most published research findigns are false. This paper defined bias in terms of design, analysis, and presentation factors with a focus on issues with p-values and statistical power. Since then there have been many replication attempts of influential (and lesser influential) papers. Strikingly, Open Science Collaboration (2015) reports 100 psychological studies run by 270 collaborators. They reported significant effects in only 36% of replications, with 47% of originally reported effects falling within 95% CIs of the replication effect. In essence: fewer significant findings and smaller effect sizes were found in replication studies compared to the original 100 studies Figure 2.
2 What is reproducibility?
- one piece of the Open Science pie
- generating the same results with the same data and analysis scripts
- seems obvious, but requires organisation and forethought before and during data collection/analysis
- bare minimum: share the code and the data (Laurinavichyute et al., 2022)
2.1 Reproducibility vs. replication
- the two terms have been used interchangably in the past (e.g., in the title of Open Science Collaboration, 2015)
- we’ll define them as follows (and this is becoming the standard distinction, imo)
Reproducibility
- re-analysing the same data using (ideally) the same scripts, software, etc
- aim: produce the same results (means, model estimates, etc.)
- why: tests for errors, coding mistakes, biases, etc.
Replication
- re-running a previous experiment, ideally with the same materials, set-up, etc.
- ideally the same analysis workflow as the original study (i.e., like reproducing the analyses but with new data)
- aim: test whether results are replicated with new data in terms of direction and magnitude
- in short:
- reproducibility = re-analysis of the same data
- replication = collection of new data
3 Why implement reproducibility in my workflow?
- firstly: the help future you (or collaborators/other researchers)!
- you may return to your analyses tomorrow, next month, or next year
- to ensure robustness and to document your steps
- ‘researcher degrees of freedom’ and the ‘garden of forking paths’: there’s more than one way to analyse a certain dataset
- we can try to plan ahead in detail (e.g., pre-reigster your analysis plan), but there will always be decisions made that were not foreseen
- lastly: it makes your life much easier and streamlines your workflow
4 How to implement reproducibility?
- not exactly straightforward
- there are degrees of reproducibility
- the rest of our time will be spent on this topic
- sharing code and data is a first step
- think of the FAIR principles of data sharing
- apply them to sharing analyses as well
4.1 Practice FAIR principles
- guidelines for sharing digital resources
- refers broadly to data, but we’ll consider it in terms of analyses

- findable and accesssible refer to where materials are stored
- in findable repositories
- that are accessible, i.e., do not require an account
- interoperable and reusable emphasise the format of data (and code)
- the importance of future use
- and use beyond your precise computational environment
4.2 Code review
- a great way to test the FAIR principles
- code review!
- i.e., have a colleague try to access your data/run your code
- either via an online repository
- or send them your project folder
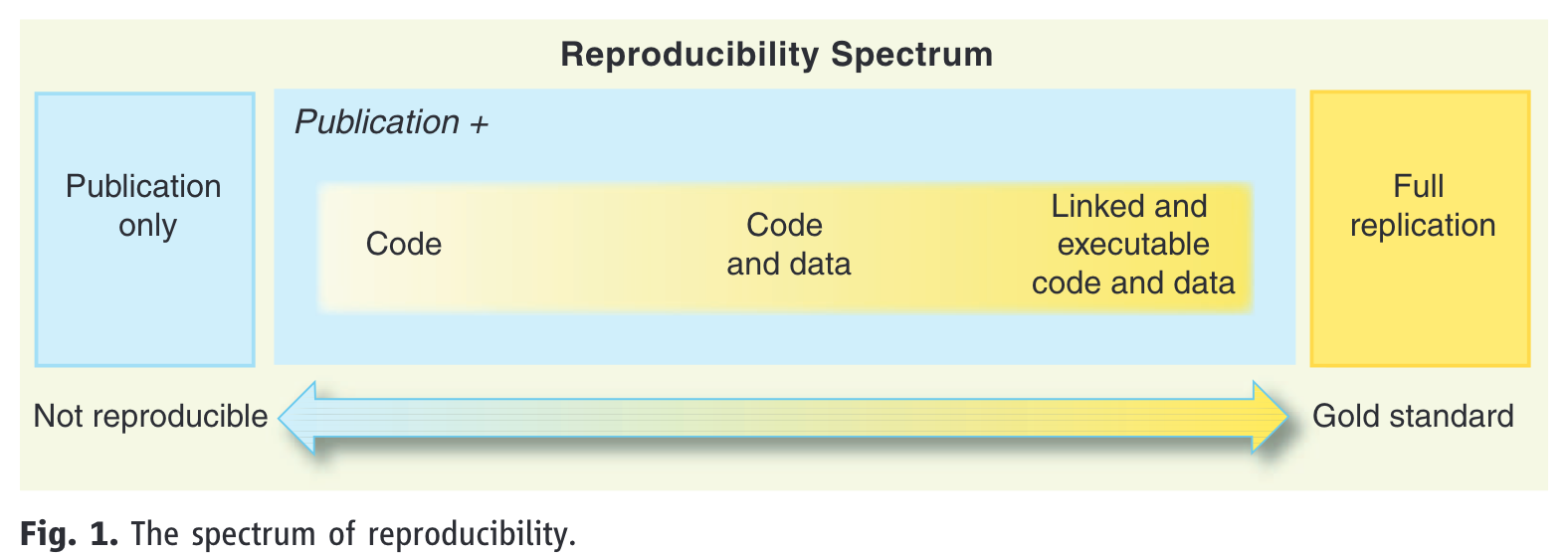
5 The reproducibility spectrum
- reproducibility is on a continuum, referred to as the reproducibility spectrum in Peng (2011) (Figure 4)
- linked means “all data, metadata, and code [is] stored and linked with each other and with corresponding publications” (Peng, 2011, p. 1227)
- executable is not explained, and is more difficult to guarantee long-term as it depends on software versions
- but at minimum we can assume it refers to code running on someone else’s machine
5.1 Data and code availability
- “data available upon (reasonable) request”
- generally not true
- data was not available in 68% of the most cited psychology studies (2006-2016) (Hardwicke & Ioannidis, 2018)
- a further 18% were available with restrictions
- only 11% available without restriction
- data alone is not sufficient
- ‘Data Analysis’ sections are rarely exhaustive/unambiguous
- very difficult to re-create analyses without code
- e.g., is data trimming explicitly defined?
- this will even affect descriptive statistics
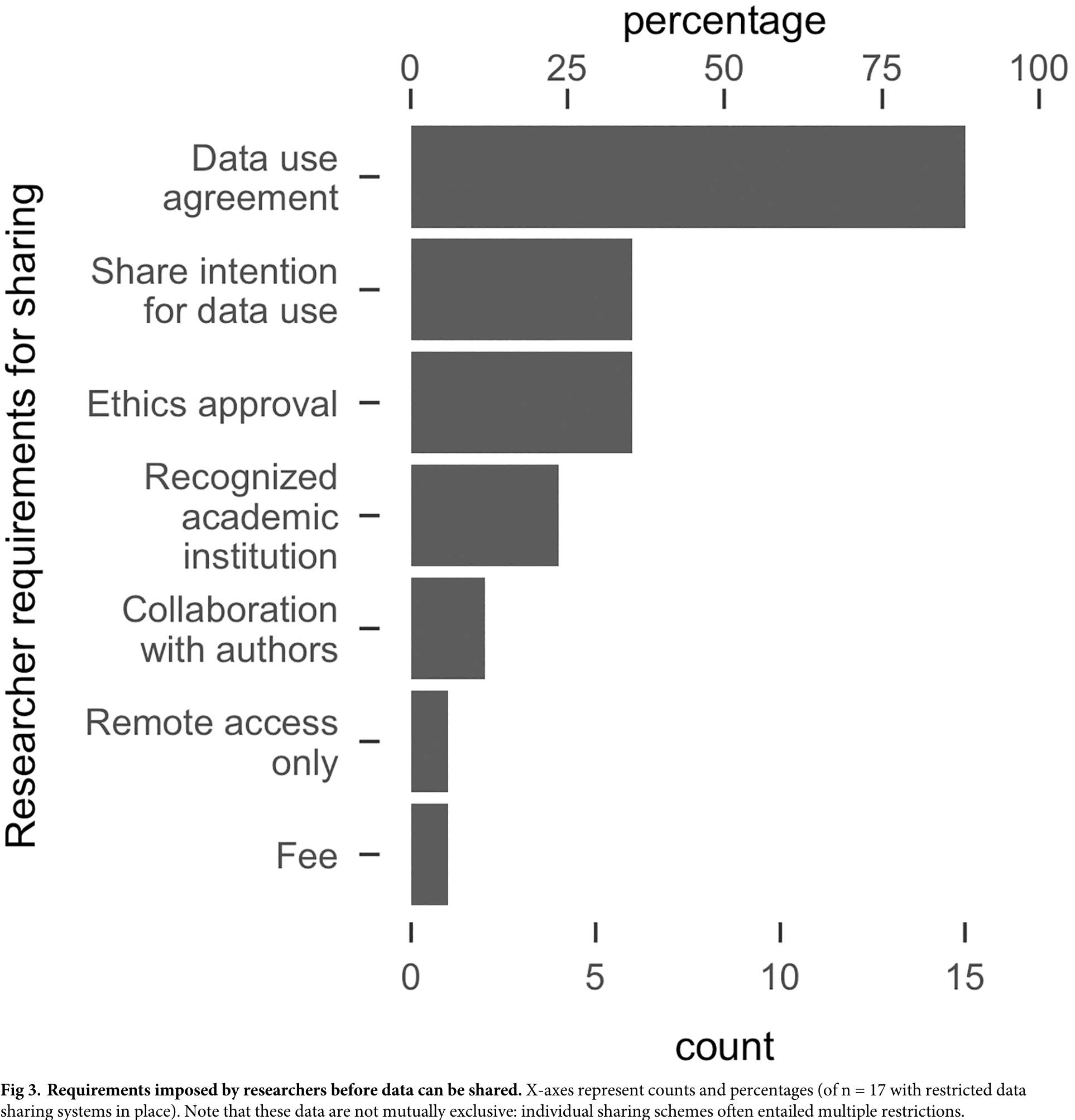
5.3 Data and code \(\neq\) Reproducibility
even including code does not guarantee reproducibility
access to data and code do not mean analyses are reproducible
what can go wrong? Examples from Laurinavichyute et al. (2022)
- Data problems
- inaccessible data
- incomplete data (e.g., 2/3 experiments)
- Code problems
- incomplete code
- error messages
- code rot: outdated syntax or environment
- proprietary software
- Documentation problems
- data difficult to interpret
- no README file/data dictionary
- unclear folder/file/variable naming convention
- manuscript contradicts code
- Unclear terms of use
- no licence specification
6 Reproducibility rates of published works
- rates of reproducibility vary across fields (see Bochynska et al., 2023 for a review)
- open access: 25-65%
- data and analyses sharing: 11-33%
- pre-registrations: 0-3%
6.1 Reproducibility rates in linguistic research
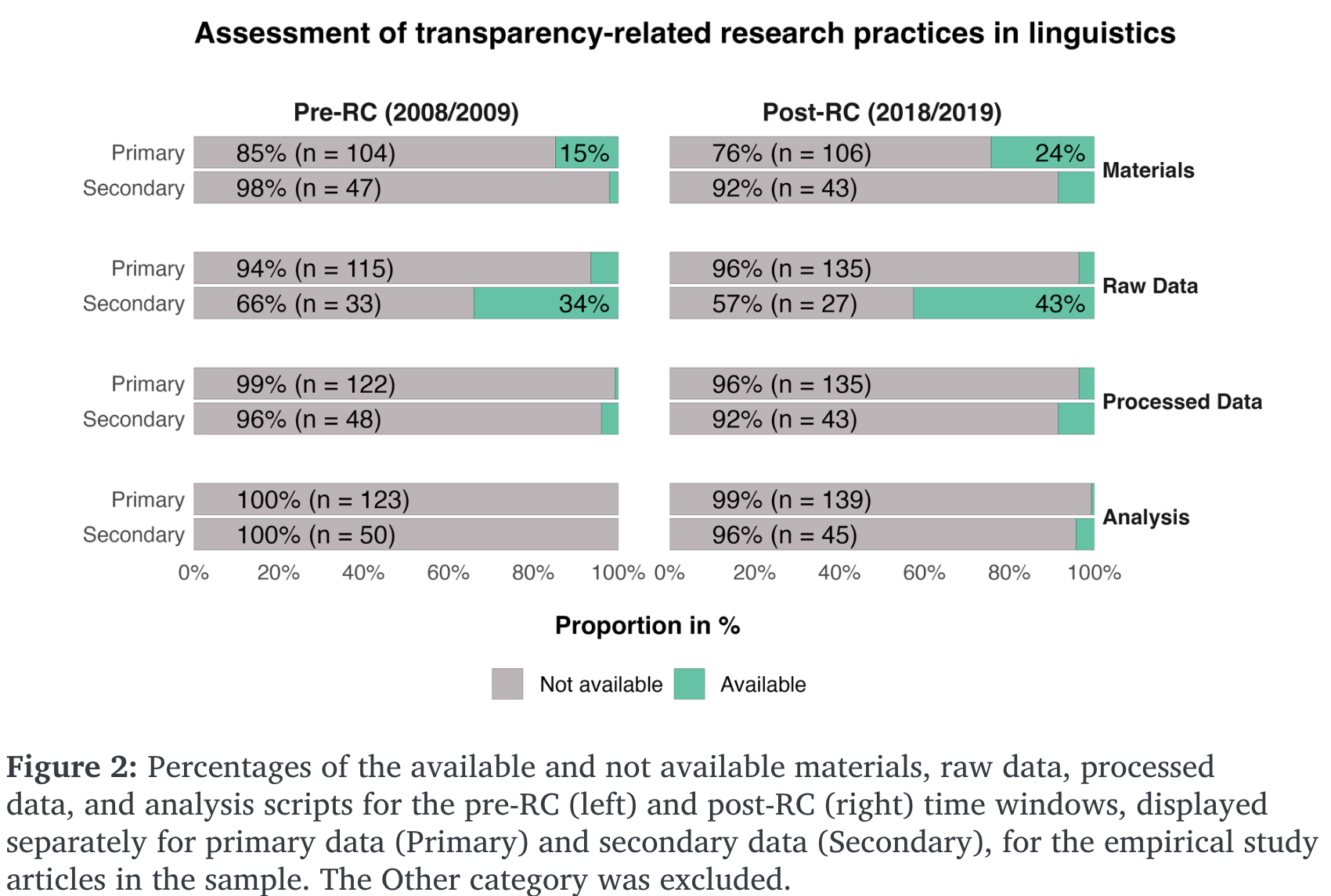
- meta-analysis of 519 randomly sampled articles from various linguistic journales
- pre- and post-reproducibility crisis (2008/9, 2018/19) (Bochynska et al., 2023)
- differentiated between primary (collected for study) and secondary (pre-existing) data
- reported a post-RC increase in shared materials, data, and analyses
- but still low rates of each
- higher rates of secondary data sharing, presumably due to publicly available corpora
- data shared more often than analyses, pre- and post-RC
6.2 Journal of Memory and Language
- meta-analysis of articles from JML (Laurinavichyute et al., 2022)
- before and after an Open Science Policy was introduced in 2019
- code and data availability improved
- but reproducibility rate ranged from 34-56%, depending on criteria
- higher rates compared to field-wide meta-analysis (Bochynska et al., 2023)
7 Steps we’ll take
- Open source software:
- R, an open source statistical programming language
- in RStudio, an IDE (integrated developer environment)
- with R Projects
- Project-oriented workflow:
- establish folder structure
- and file/variable naming conventions
- use project-relative filepaths with the
herepackage - establish and maintain project-relative package library with
renv(time permitting)
- Practice literate programming:
- writing clean, commented, linear code
- in dynamic reports (e.g., Quarto, R markdown)
- practice modularity, i.e., 1 script = 1 purpose
- Sharing and checking our code
- uploading our code and data to an OSF repository
- conducting a code review
Learning objectives 🏁
Today we learned…
- what ‘reproducibility’ is and why/how to practice it ✅
- FAIR principles for sharing our code and data ✅
- concepts for building a reproducible workflow ✅