.Rproj
Creating a project-oriented workflow in R
Daniela Palleschi
Humboldt-Universität zu Berlin
Thu Aug 22, 2024
Wed Aug 21, 2024
Today we will…
here package4.4.0, “Puppy Cup”R.version2023.12.1.402, “Ocean Storm”.Rproj file in a project folderSoSe2024 or MastersarbeitFile > New Project > New Directory > New Project > [Directory name] > Create ProjectCreate a new RProject for this workshop
File > New Project > New Directory > New Project > [Directory name] > Create Project.Rproj file and double-clickProject (None) drop-down (top right)
.Rproj
File > New File > Markdown File (not R Markdown!)
# for headings, *italics*, **bold**)README.md in your project directory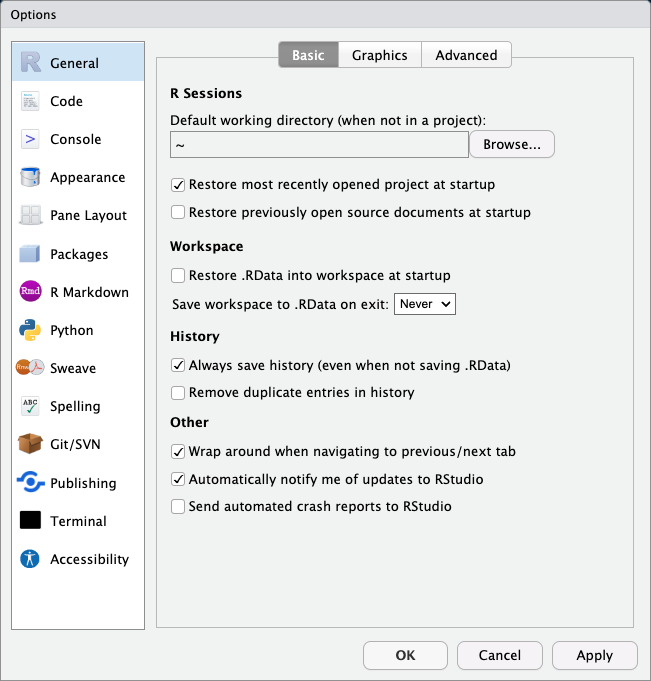
Tools > Global Options
Change your Global Options so that
data: containing your dataset(s)scripts (or analyses, etc.): containing any analysis scriptsmanuscript: containing any write-ups of your resultsmaterials: containing relevant experiment materials (e.g., stimuli)data and scripts)data/raw sub-folderprocessed or tidy)scripts/Create a new Quarto script:
File > New File > Quarto DocumentUse Visual Editor boxCreatescripts/ folder: File > Save as...readr::read_csv()here-packagehere package (Müller, 2020) enables file referencing
setwd()
setwd()If the first line of your R script is
setwd("C:\Users\jenny\path\that\only\I\have")I will come into your office and SET YOUR COMPUTER ON FIRE🔥.
setwd() depends on your entire machine’s folder structuresetwd() breaks when you
here()here
Load the dataset using here
here (e.g., install.packages("here"))here at the beginning of your package
here:: before calling a functionhere() function to load in your datasummary(), names(), etc.)here::here()here functionhere function without loading the packagedf_
df stands for dataframefit_ for models, fig_ for figures, sum_ for summaries, tbl_ for tables, etc.names(), summary(), dplyr::glimpse(), whatever you typically do)Today we learned…
here ✅---
title: "RProjects"
subtitle: "Creating a project-oriented workflow in R"
author: "Daniela Palleschi"
institute: Humboldt-Universität zu Berlin
lang: en
date: 2024-08-22
date-format: "ddd MMM D, YYYY"
date-modified: last-modified
language:
title-block-published: "Workshop Day 2"
title-block-modified: "Last Modified"
format:
html:
output-file: rprojects.html
number-sections: false
toc: true
code-overflow: wrap
code-tools: true
embed-resources: false
pdf:
output-file: rprojects.pdf
toc: true
number-sections: false
colorlinks: true
code-overflow: wrap
revealjs:
footer: "SSOL 2024"
output-file: rprojects_slides.html
code-overflow: wrap
theme: [dark]
width: 1600
height: 900
# chalkboard:
# src: chalkboard.json
progress: true
scrollable: true
# smaller: true
slide-number: c/t
code-link: true
incremental: true
# number-sections: true
toc: false
toc-depth: 2
toc-title: 'Overview'
navigation-mode: linear
controls-layout: bottom-right
fig-cap-location: top
font-size: 0.6em
slide-level: 4
embed-resources: false
fig-align: center
fig-dpi: 300
editor_options:
chunk_output_type: console
bibliography: references.bib
csl: ../../../apa.csl
execute:
echo: false
---
```{r}
#| echo: false
#| eval: false
rbbt::bbt_update_bib(here::here("slides", "day2", "rprojects", "rprojects.qmd"))
```
# Learning Objectives {.unnumbered .unlisted}
Today we will...
- learn about project-oriented workflows
- create an RProject
- use project-relative filepaths with the `here` package
# Installation requirements
- required installations/recent versions of:
- R
- version `4.4.0`, "Puppy Cup"
- check current version with `R.version`
- download/update: <https://cran.r-project.org/bin/macosx/>
- RStudio
- version `2023.12.1.402`, "Ocean Storm"
- Help \> Check for updates
- new install: <https://posit.co/download/rstudio-desktop/>
# Project-oriented workflow
1. Folder structure:
+ keeping everything related to a project in one place
+ i.e., contained in a single folder, with subfolders as needed
2. Project-relative working directory
+ the project folder should act as your working directory
+ all file paths should be relative to this folder
## Folder structure
- a core computer literacy skill
+ keep your Desktop as empty as possible
+ have a sensible folder structure
+ avoid mixing subfolders and files
+ i.e., if a folder contains subfolders, ideally it should not contain files
# RProjects
- in data analysis, using an IDE is beneficial
+ e.g., RStudio
- most IDEs have their own implementation of a Project
- in RStudio, this is the RProject
+ creates a `.Rproj` file in a project folder
+ stores project settings
- you can have several RProjects open simultaneously
+ and run several scripts across projects simultaneously
- most importantly, RProjects (can) centralise a specific project's workflow and file path
- to read more about R Projects, check out [Section 6.2: Projects](https://r4ds.hadley.nz/workflow-scripts.html#projects) from @wickham_r_2023 [or [Ch. 8 - Workflow: Projects](https://r4ds.had.co.nz/workflow-projects.html) in @wickham_r_2016]
## Creating a new Project
- when?
+ whenever you're starting a new course or project which will use R
- why?
+ to keep all the relavent materials in one place
- where?
+ somewhere that makes sense, e.g., a folder called `SoSe2024` or `Mastersarbeit`
- how?
+ `File > New Project > New Directory > New Project > [Directory name] > Create Project`
### {.unnumbered .unlisted}
::: {.callout-tip}
# New RProject
Create a new RProject for this workshop
+ `File > New Project > New Directory > New Project > [Directory name] > Create Project`
+ make sure you choose a sensible location
:::
## Opening a Project
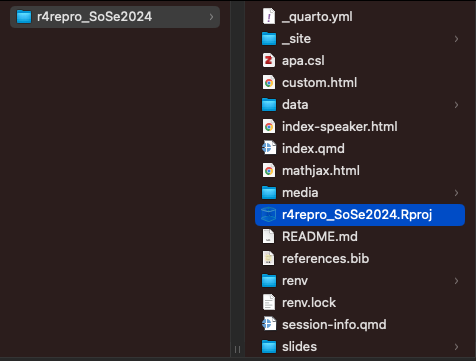
- to open a project, locate its `.Rproj` file and double-click
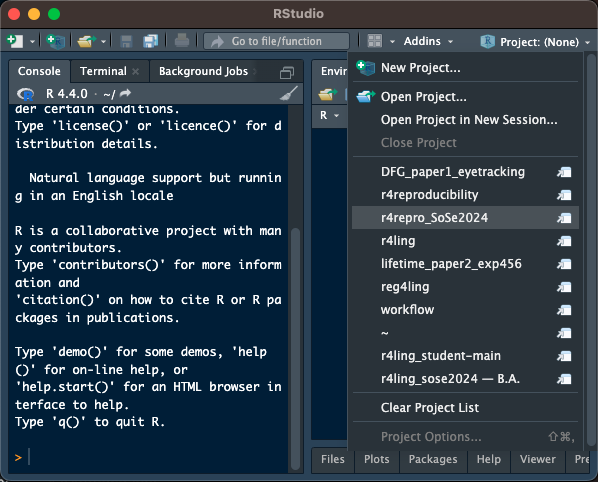
- or if you're already in RStudio, you can use the `Project (None)` drop-down (top right)
:::: {.columns}
::: {.column width="50%"}
```{r}
#| label: fig-click-open
#| fig-cap: Double-click `.Rproj`
#| out-width: "80%"
knitr::include_graphics(here::here("media", "rstudio_click_open.png"))
```
:::
::: {.column width="50%"}
```{r}
#| label: fig-project-open
#| fig-cap: Open from RStudio
knitr::include_graphics(here::here("media", "rstudio_project_open.png"))
```
:::
::::
## Adding a README file
- `File > New File > Markdown File` (*not* R Markdown!)
+ add some text describing the purpose of this project
+ include your name, the date
+ use Markdown formatting (e.g., `#` for headings, `*italics*`, `**bold**`)
- save as `README.md` in your project directory
## Global RStudio options
:::: {.columns}
::: {.column width="50%"}
```{r}
#| label: fig-rstudio-settings
#| fig-cap: RStudio settings for reproducibility
knitr::include_graphics(here::here("media", "RStudio_global-options.png"))
```
:::
::: {.column width="50%"}
- `Tools > Global Options`
+ **Workspace**: Restore .RData into workspace at startup: NO
+ Save workspace to .RData on exit: Never
- this will ensure that you are always starting with a clean slate
+ and that your code is not dependent on some pacakge or object you created in another session
- this is also how RMarkdown and Quarto scripts run
+ they start with an empty environment and run the script linearly
:::
::::
## {.unnumbered .unlisted}
::: {.callout-tip}
## Global settings
Change your Global Options so that
+ **Workspace**: Restore .RData into workspace at startup: NO
+ Save workspace to .RData on exit: Never
:::
## Identifying your RProject
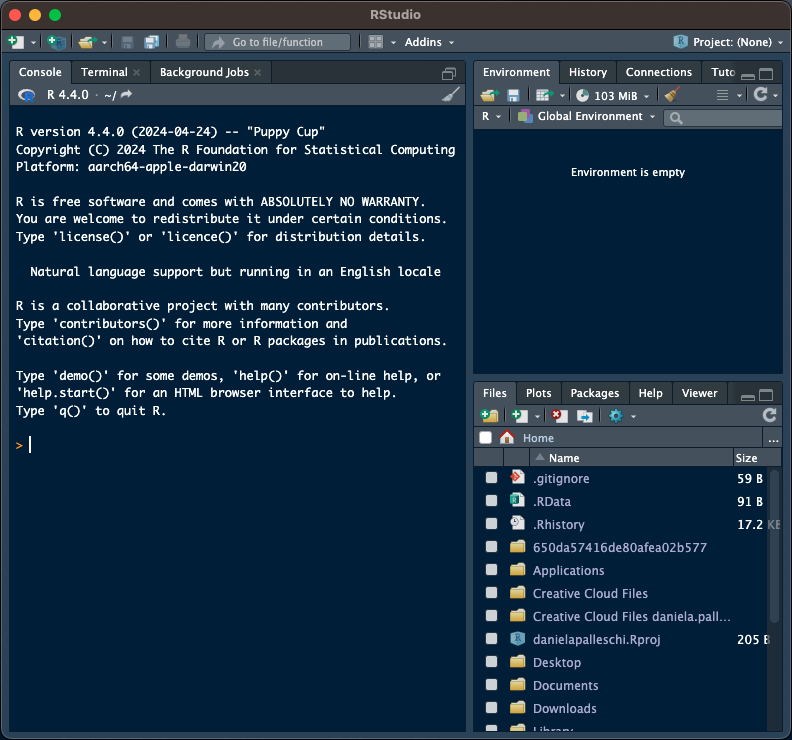
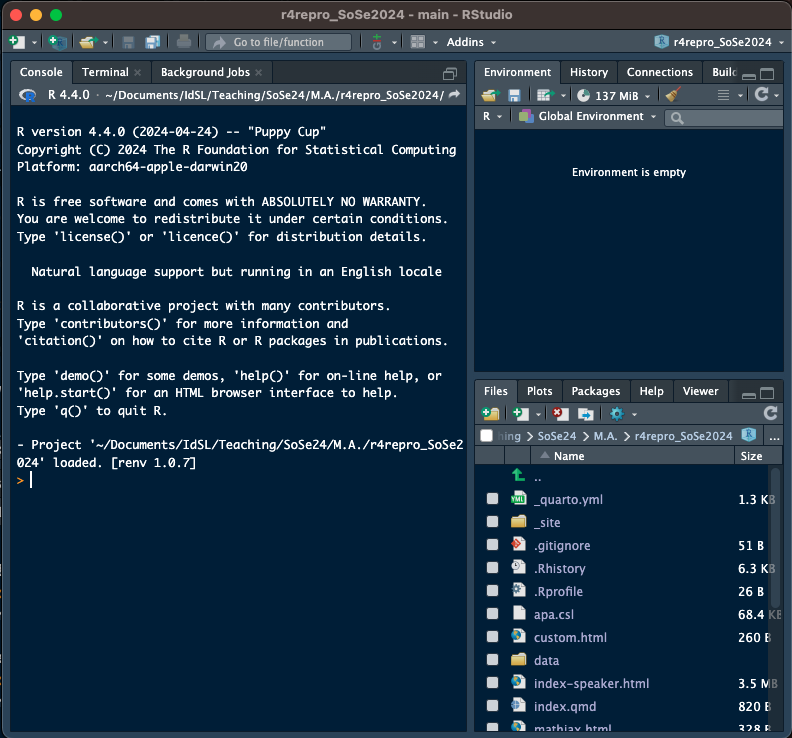
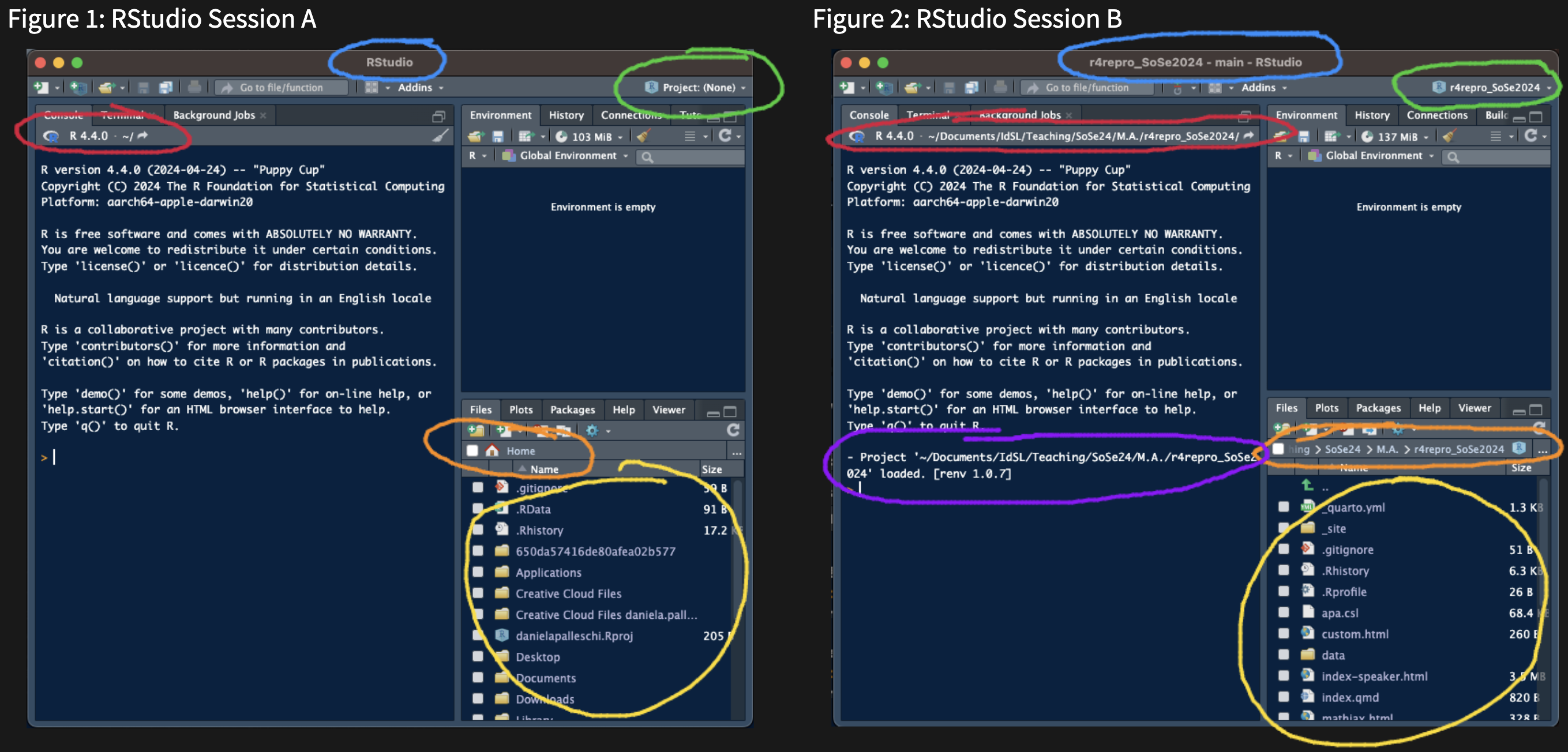
- there are a ways to check which (if any) RProject you're in
+ there are 6 differences between xyzfig-noproject and xyzfig-project
+ which is in an RProject session?
::: {.panel-tabset}
### Spot the differences
:::: {.columns}
::: {.column width="45%"}
```{r}
#| label: fig-noproject
#| fig-cap: RStudio Session A
knitr::include_graphics(here::here("media", "rstudio_noproject.png"))
```
:::
::: {.column width="5%"}
:::
::: {.column width="45%"}
```{r}
#| label: fig-project
#| fig-cap: RStudio Session B
knitr::include_graphics(here::here("media", "rstudio_project.png"))
```
:::
::::
### Show the differences
```{r}
knitr::include_graphics(here::here("media", "rproject_spot-the-diffs.png"))
```
:::
# Folder structure
- some folders you'll typically want to have:
+ `data`: containing your dataset(s)
+ `scripts` (or `analyses`, etc.): containing any analysis scripts
+ `manuscript`: containing any write-ups of your results
+ `materials`: containing relevant experiment materials (e.g., stimuli)
- let's just create the first 2 (`data` and `scripts`)
### `data/`
- do you have "raw", i.e., pre-processed data?
+ if so, you might want to create a `raw` sub-folder
+ and any other relevant sub-folders (e.g., `processed` or `tidy`)
- download [the dataset](https://osf.io/ushpw) from the workshop repo [from @chromy_number_2023]
+ *or*, move a dataset of your own to this folder
### `scripts/`
- try to create a single script for each "product"
+ e.g., anonymised data, 'cleaned' data, data exploration, visualisation, analyses, etc.
- you can create sub-folders as the project develops and move scripts around
+ for now, let's create a new script to take a look at our data
### {.unnumbered .unlisted}
::: {.callout-tip}
## New script
Create a new Quarto script:
1. `File > New File > Quarto Document`
3. Add a title
2. Uncheck the `Use Visual Editor` box
4. Click `Create`
5. Save it in your `scripts/` folder: `File > Save as...`
:::
### Load in the data
- load in the data however you normally would
+ e.g., `readr::read_csv()`
# `here`-package
- `here` package [@here-package] enables file referencing
+ avoids the use of `setwd()`
```{r}
#| label: fig-here
#| fig-cap: Illustration by [Allison Horst](https://github.com/allisonhorst)
knitr::include_graphics(here::here("media", "Horst_here.png"))
```
## The problem with `setwd()`
> If the first line of your R script is
>
> `setwd("C:\Users\jenny\path\that\only\I\have")`
>
> I will come into your office and SET YOUR COMPUTER ON FIRE🔥.
--- [Jenny Bryan](https://x.com/hadleywickham/status/940021008764846080)
- `setwd()` depends on your entire machine's folder structure
- `setwd()` breaks when you
+ send your project folder to a collaborator
+ make your analyses open
+ change the location of your project folder
- using slashes is also dependent on your operating system
## The benefit of `here()`
- uses the top-level directory of your project as the working directory
- can separate folder names with a comma
## {.unlisted .unnumbered}
::: {.callout-tip}
# `here`
Load the dataset using `here`
1. Install `here` (e.g., `install.packages("here")`)
2. Load `here` at the beginning of your package
+ or use `here::` before calling a function
3. Use the `here()` function to load in your data
4. Inspect the dataset however you usually would (e.g., `summary()`, `names()`, etc.)
4. Save your script
:::
## `here::here()`
- install package
```{r filename = "In the Console"}
#| eval: false
#| echo: true
install.packages("here")
```
- load package and call the `here` function
```{r}
#| eval: false
#| echo: true
# load package
library(here)
# read in data
df_data <- read.csv(here("data", "data_lifetime_pilot.csv"))
```
- or directly call the `here` function without loading the package
```{r}
#| eval: false
#| echo: true
# read in data without loading here
df_data <- read.csv(here::here("data", "data_lifetime_pilot.csv"))
```
- note that I stored the data with the prefix `df_`
+ `df` stands for dataframe
- I recommend using object-type defining prefixes for all objects in your Environment
+ e.g., `fit_` for models, `fig_` for figures, `sum_` for summaries, `tbl_` for tables, etc.
## {.unlisted .unnumbered}
::: {.callout-tip}
# Reproduce your analysis
1. Perform some data exploration (e.g., with `names()`, `summary()`, `dplyr::glimpse()`, whatever you typically do)
1. Save your script, then close RStudio/your Rproject.
2. Re-open the project. Can you re-run the script?
:::
# Learning objectives 🏁 {.unnumbered .unlisted .uncounted}
Today we learned...
- learn about project-oriented workflows ✅
- create an RProject ✅
- establish a self-contained project environment with `here` ✅
# References {.unlisted .unnumbered visibility="uncounted"}
---
nocite: |
@bryan_what_nodate-1
@bryan_chapter_nodate
@noauthor_using_2024
---
::: {#refs custom-style="Bibliography"}
:::